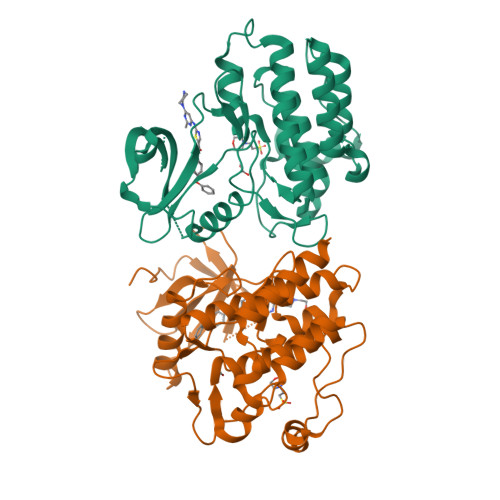
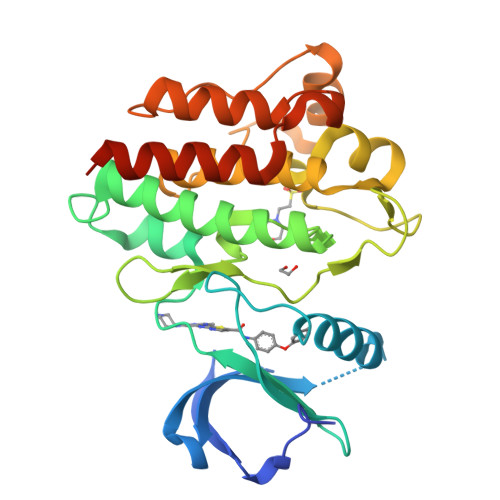
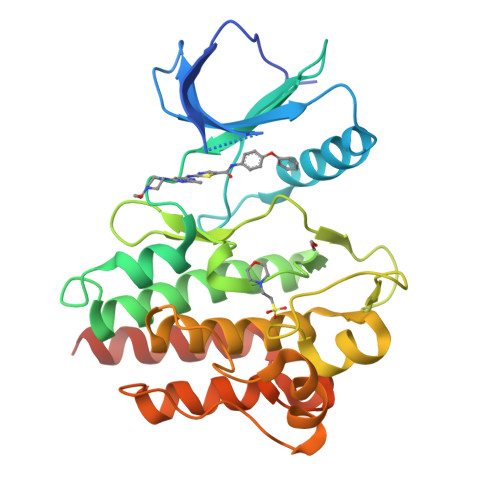
Conformation-Selective Analogues of Dasatinib Reveal Insight into Kinase Inhibitor Binding and Selectivity.
Kwarcinski, F.E., Brandvold, K.R., Phadke, S., Beleh, O.M., Johnson, T.K., Meagher, J.L., Seeliger, M.A., Stuckey, J.A., Soellner, M.B.(2016) ACS Chem Biol 11: 1296-1304
- PubMed: 26895387
- DOI: https://doi.org/10.1021/acschembio.5b01018
- Primary Citation of Related Structures:
4YBJ, 4YBK, 4YC8 - PubMed Abstract:
In the kinase field, there are many widely held tenets about conformation-selective inhibitors that have yet to be validated using controlled experiments. We have designed, synthesized, and characterized a series of kinase inhibitor analogues of dasatinib, an FDA-approved kinase inhibitor that binds the active conformation. This inhibitor series includes two Type II inhibitors that bind the DFG-out inactive conformation and two inhibitors that bind the αC-helix-out inactive conformation. Using this series of compounds, we analyze the impact that conformation-selective inhibitors have on target binding and kinome-wide selectivity.
Organizational Affiliation:
Department of Pharmacological Sciences, Stony Brook University , Stony Brook, New York 11794, United States.